|
Leigh Brown HIV Research Group | ||
 |
|||||||||
|
|||||||||
 |
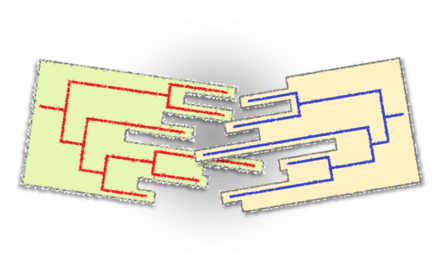
Picking and Describing HIV Clusters in Phylogenetic Trees A tool combination for the analysis of phylogenetic clusters of nucleotide sequences
Description Cluster identification strategies differ between studies and as a consequence cluster definitions vary. The Cluster Picker identifies clusters in newick-formatted trees containing thousands of sequences within a few minutes. Cut-offs for within cluster genetic distance and bootstrap support are selected by the user. Because many groups then look at the epidemiology of these clusters, the Cluster Matcher automatically links Cluster Picker output to spreadsheets of epidemiological data. Associated publication Ragonnet-Cronin M, Lycett SJ, Hodcroft E, Hue S, Fearnhill E, Dunn D, Delpech V, Leigh Brown AJ. Automated analysis of phylogenetic clusters. BMC Bioinformatics 14:317; 2013 link to journal article. Installation To run the Cluster Picker and Cluster Matcher, you will need Java 1.6.0 or higher. The jar file will run on PC, Mac, UNIX and LINUX operating systems. Source Code
https://github.com/emmahodcroft/cluster-picker-and-cluster-matcher
|
 |
|||||||